DDPMs for MRI
Diffusion-Driven Generation of Minimally Preprocessed Brain MRI
Background and Methods
Training a deep generative model for 3D medical images, such as magnetic resonance (MR) images, is a challenging task. The current state-of-the-art uses Denoising Diffusion Probabilistic Models (DDPMs), which progressively transform noise into a sample from the training distribution. As MR images are frequently acquired in 3D (or stacked 2D slices), a generative model needs 3D consistency, which means (usually) a 3D network. 3D DDPMs, however, are large, data-hungry, and slow.
To train one of these large models, we needed a very large training set of varied, but still high-quality data. We collected 38 publically available datasets and curated them for high-resolution, high-quality T$_1$-weighted brain images (see our blog post for more details).
We want our models to be useful to as many users as possible, so we avoided some of the common steps used to reduce the complexity of the data (registration, bias correction, downsampling, latent models, etc.). 3D DDPMs are big models, so tested different prediction methods to determine which would be stable and converge (relatively) quickly. This led us to train a whole suite of models using different prediction methods to generate the most realistic images possible. DDPMs use a U-Net to predict how to denoise an image, but that can be done in multiple ways. Commonly, networks will either predict the noise ($\hat{\epsilon}$) or the sample (sample, $\hat{x}_0$). But groups have also proposed methods that predict weighted or unweighted sums of the noise and clean image, called velocity ($\hat{\nu}$) prediction and flow ($\hat{\mu}$) matching, respectively. We wanted to try all of these methods to evaluate which would provide the best results.
Results
We found that in our training, noise prediction failed to converge, while sample, velocity and flow models all produced brain-like samples.
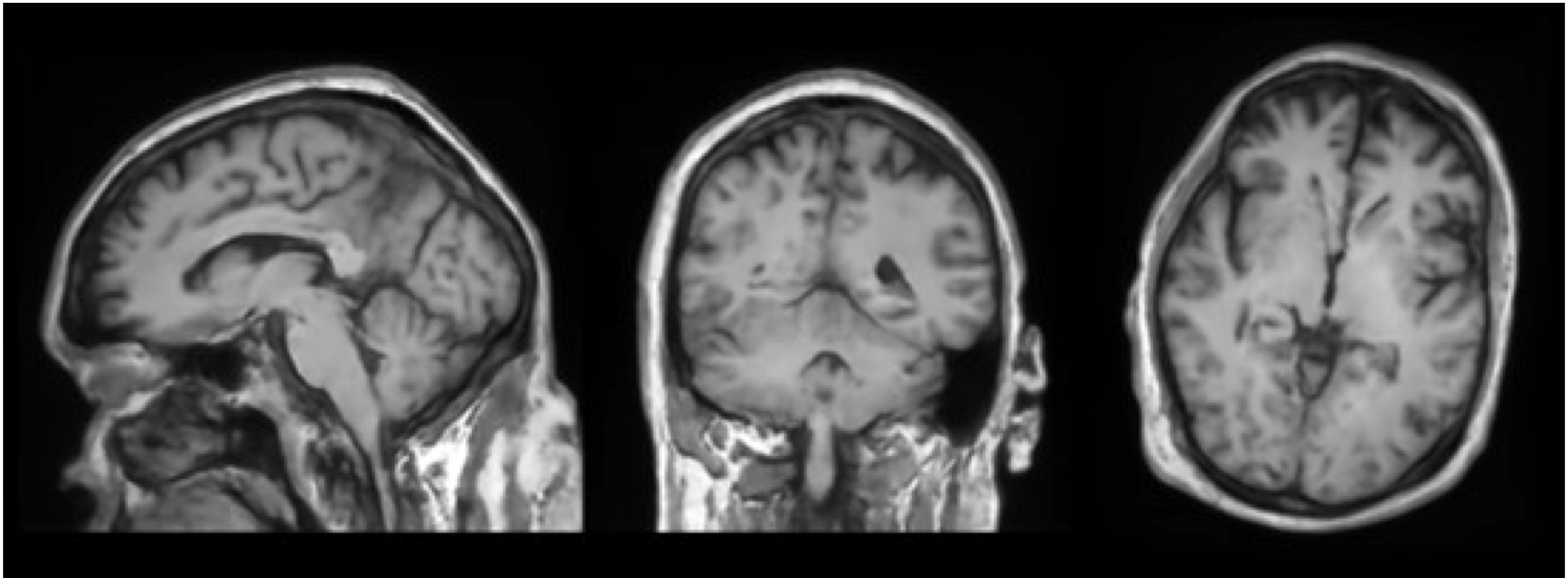
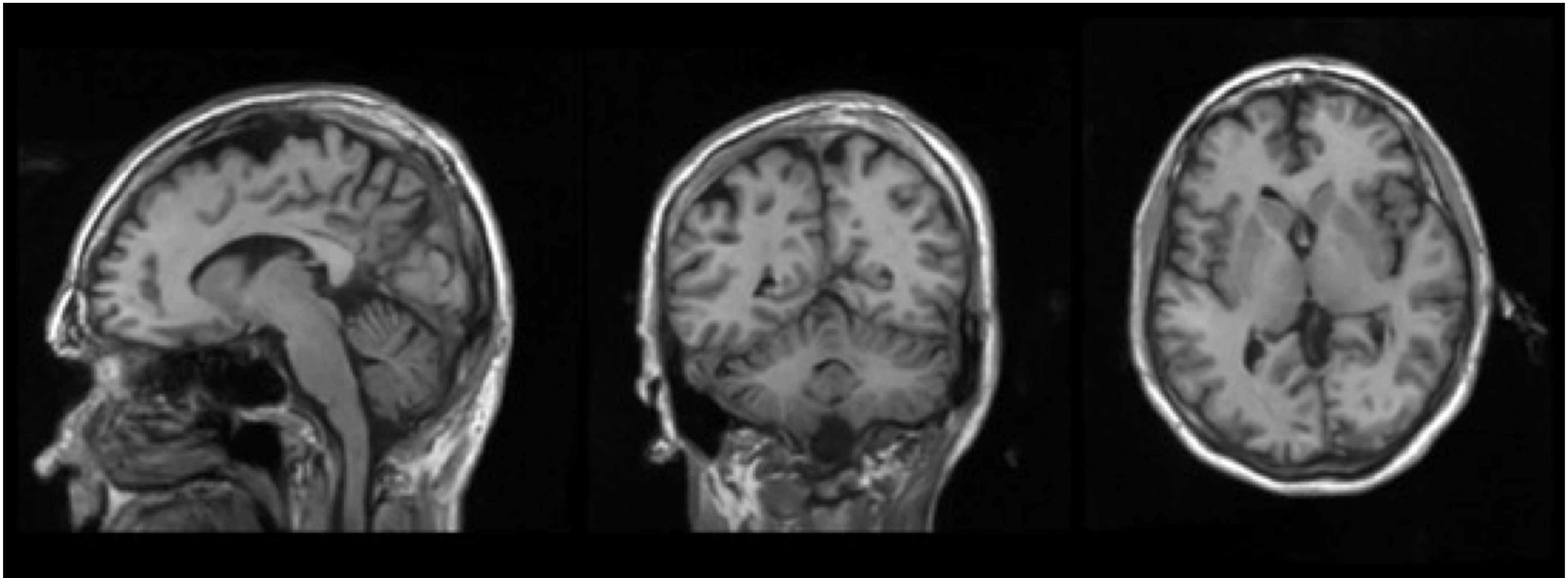
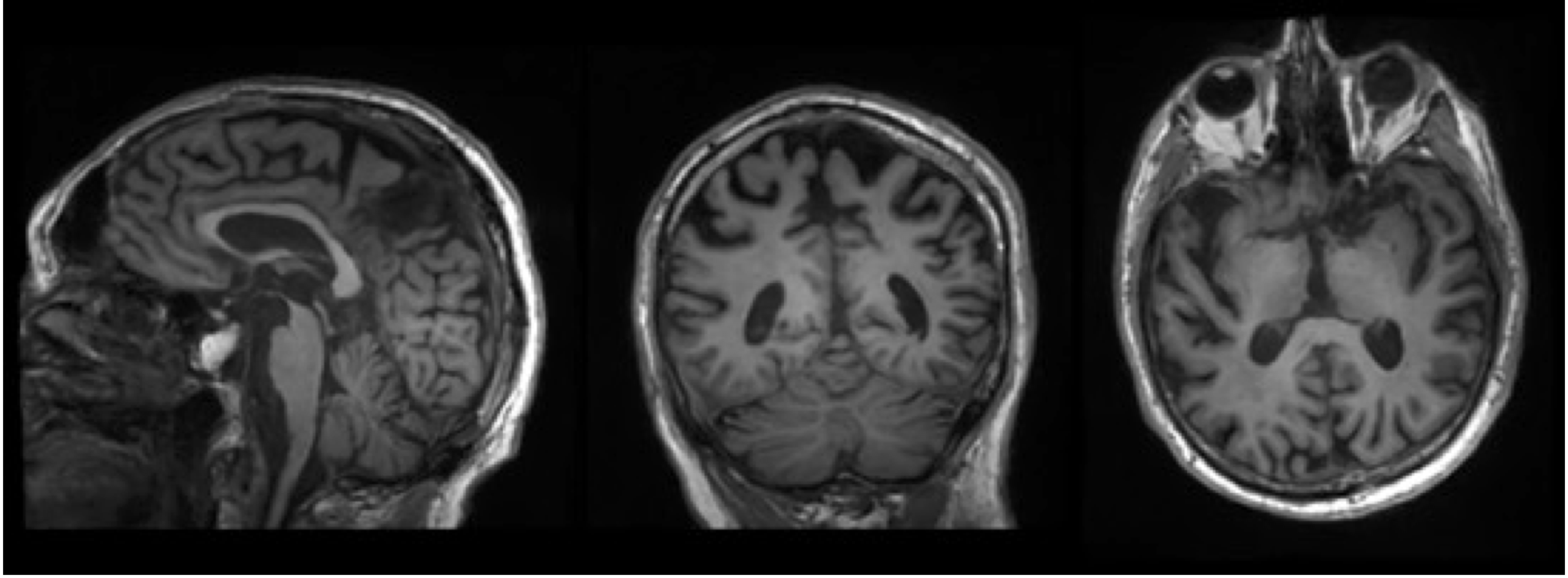
Prediction methods: top left - sample, top right - velocity, bottom - flow.
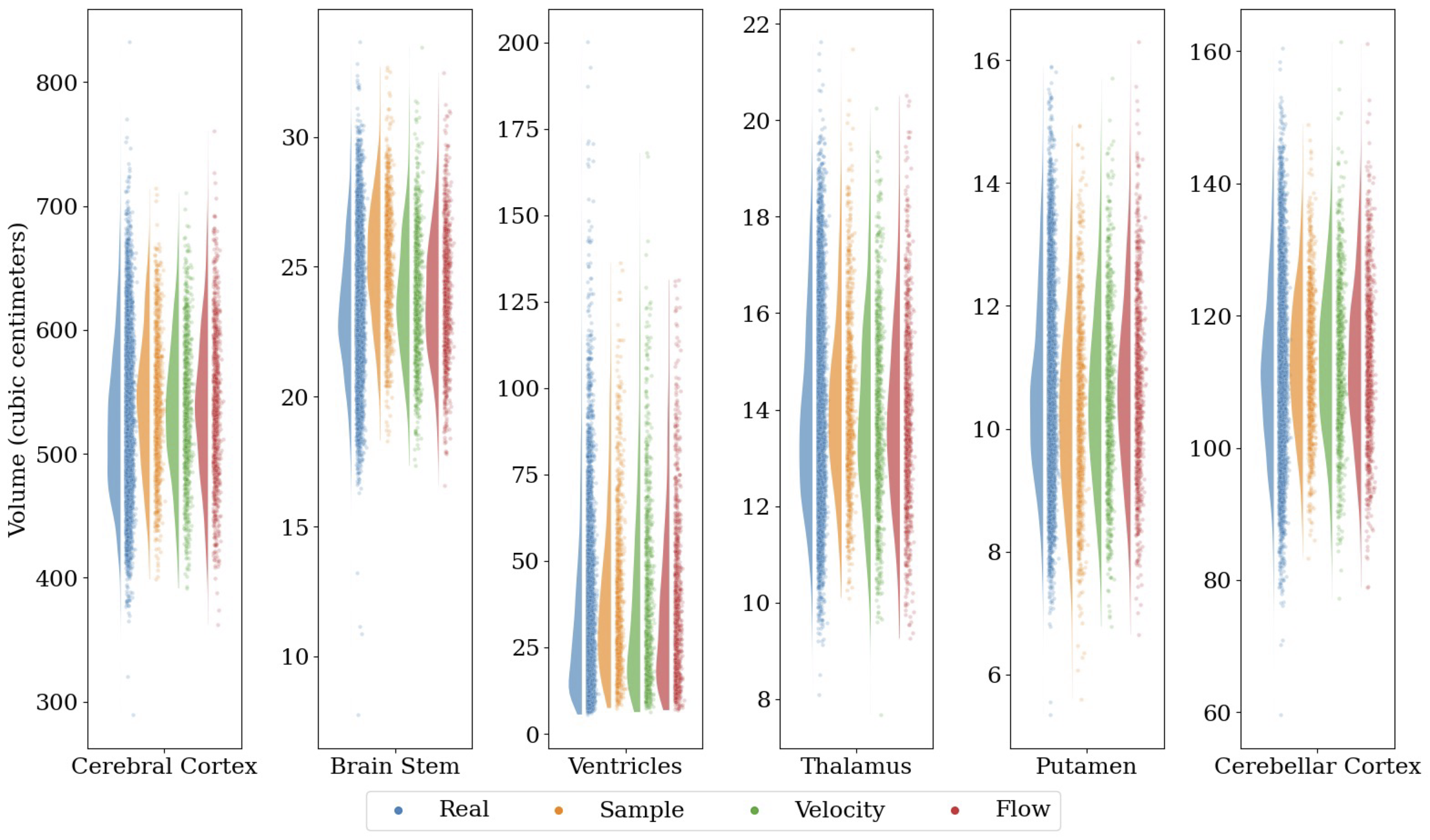
We also showed that our synthetic images (except for noise prediction outputs) can be segmented by methods like SynthSeg. Their volumes, while not exactly aligned with the testing distribution, were quite close, with velocity and flow prediction methods aligning better than sample results.
Paper and Models
Our paper is currently under-review with a preprint on the way soon.
Code (and pre-trained weights) for these models is available on Github